| Revision as of 12:33, 30 November 2019 editCobaltcigs (talk | contribs)Autopatrolled, Extended confirmed users28,629 editsm refs← Previous edit | Latest revision as of 21:44, 6 October 2024 edit undoUtini4 (talk | contribs)159 editsm no longer an orphan | ||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| {{primary sources|date=November 2021}} | |||
| {{DISPLAYTITLE:''nhaA''-II RNA motif}} | {{DISPLAYTITLE:''nhaA''-II RNA motif}} | ||
| {{Infobox rfam | {{Infobox rfam | ||
| Line 24: | Line 26: | ||
| | LocusSupplementaryData = | | LocusSupplementaryData = | ||
| }} | }} | ||
| The '''''nhaA''-II RNA motif''' is a conserved ] structure that was ].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages=10811–10823 |date=October 2017 |pmid=28977401 |pmc=5737381 |doi=10.1093/nar/gkx699 |
The '''''nhaA''-II RNA motif''' is a conserved ] structure that was ].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages=10811–10823 |date=October 2017 |pmid=28977401 |pmc=5737381 |doi=10.1093/nar/gkx699 }}</ref> | ||
| ''nhaA''-II motifs are found in ]. | ''nhaA''-II motifs are found in ]. | ||
Latest revision as of 21:44, 6 October 2024
| This article relies excessively on references to primary sources. Please improve this article by adding secondary or tertiary sources. Find sources: "NhaA-II RNA motif" – news · newspapers · books · scholar · JSTOR (November 2021) (Learn how and when to remove this message) |
| nhaA-II | |
|---|---|
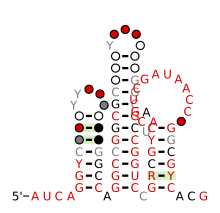 Consensus secondary structure and sequence conservation of nhaA-II RNA Consensus secondary structure and sequence conservation of nhaA-II RNA | |
| Identifiers | |
| Symbol | nhaA-II |
| Rfam | RF03038 |
| Other data | |
| RNA type | Cis-reg |
| SO | SO:0005836 |
| PDB structures | PDBe |
The nhaA-II RNA motif is a conserved RNA structure that was discovered by bioinformatics. nhaA-II motifs are found in Caulobacterales.
nhaA-II motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. nhaA-I RNAs typically occur upstream of genes that encode exchangers of sodium ions and protons. More rarely, they also exist upstream of methyltransferases that use S-adenosylmethionine as a donor.
References
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.