| Revision as of 03:55, 7 December 2018 editDouble sharp (talk | contribs)Autopatrolled, Extended confirmed users, Page movers, File movers, Pending changes reviewers102,090 editsNo edit summary← Previous edit | Revision as of 14:00, 24 January 2019 edit undoAntonipetrov (talk | contribs)327 editsm Add Rfam infoboxNext edit → | ||
| Line 1: | Line 1: | ||
| {{Infobox rfam | |||
| | Name = nhaA-II | |||
| | image = RF03038.svg | |||
| | width = | |||
| | caption = Consensus ] and ] of nhaA-II RNA | |||
| | Symbol = nhaA-II | |||
| | AltSymbols = | |||
| | Rfam = RF03038 | |||
| | miRBase = | |||
| | miRBase_family = | |||
| | RNA_type = ] | |||
| | Tax_domain = | |||
| | GO = | |||
| | SO = {{SO|0005836}} | |||
| | CAS_number = | |||
| | EntrezGene = | |||
| | HGNCid = | |||
| | OMIM = | |||
| | RefSeq = | |||
| | Chromosome = | |||
| | Arm = | |||
| | Band = | |||
| | LocusSupplementaryData = | |||
| }} | |||
| {{DISPLAYTITLE:''nhaA''-II RNA motif}} | {{DISPLAYTITLE:''nhaA''-II RNA motif}} | ||
| The '''''nhaA''-II RNA motif''' is a conserved ] structure that was ].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages=10811–10823 |date=October 2017 |pmid=28977401 |pmc=5737381 |doi=10.1093/nar/gkx699 |url=}}</ref> | The '''''nhaA''-II RNA motif''' is a conserved ] structure that was ].<ref name="Weinberg2017b">{{cite journal |vauthors=Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR |title=Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions |journal=Nucleic Acids Res. |volume=45 |issue=18 |pages=10811–10823 |date=October 2017 |pmid=28977401 |pmc=5737381 |doi=10.1093/nar/gkx699 |url=}}</ref> | ||
Revision as of 14:00, 24 January 2019
RNA family| nhaA-II | |
|---|---|
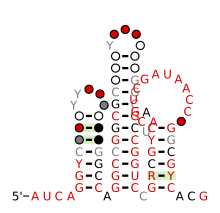 Consensus secondary structure and sequence conservation of nhaA-II RNA Consensus secondary structure and sequence conservation of nhaA-II RNA | |
| Identifiers | |
| Symbol | nhaA-II |
| Rfam | RF03038 |
| Other data | |
| RNA type | Cis-reg |
| SO | SO:0005836 |
| PDB structures | PDBe |
The nhaA-II RNA motif is a conserved RNA structure that was discovered by bioinformatics.
nhaA-II motifs are found in Caulobacterales.
nhaA-II motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes. nhaA-I RNAs typically occur upstream of genes that encode exchangers of sodium ions and protons. More rarely, they also exist upstream of methyltransferases that use S-adenosylmethionine as a donor.
References
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR (October 2017). "Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions". Nucleic Acids Res. 45 (18): 10811–10823. doi:10.1093/nar/gkx699. PMC 5737381. PMID 28977401.