This is an old revision of this page, as edited by Calthinus (talk | contribs) at 18:38, 18 March 2020 (→Infection: moved from 2019-20 coronavirus pandemic per suggestion by Robertpedley). The present address (URL) is a permanent link to this revision, which may differ significantly from the current revision.
Revision as of 18:38, 18 March 2020 by Calthinus (talk | contribs) (→Infection: moved from 2019-20 coronavirus pandemic per suggestion by Robertpedley)(diff) ← Previous revision | Latest revision (diff) | Newer revision → (diff) Virus strain causing coronavirus disease 2019 This article is about the virus strain that causes COVID-19. For the strain that causes SARS, see Severe acute respiratory syndrome coronavirus. For the species to which both strains belong, see Severe acute respiratory syndrome-related coronavirus.Template:Use Commonwealth English
| Severe acute respiratory syndrome coronavirus 2 | |
|---|---|

| |
| Electron micrograph of SARS-CoV-2 virions with visible coronae | |
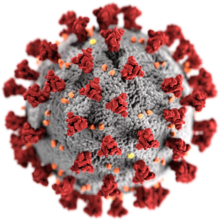
| |
| Illustration of a SARS-CoV-2 virion | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Sarbecovirus |
| Species: | Severe acute respiratory syndrome-related coronavirus |
| Strain: | Severe acute respiratory syndrome coronavirus 2 |
| Synonyms | |
|
2019-nCoV, HCoV-19 | |
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously known by the provisional name 2019 novel coronavirus (2019-nCoV), is a positive-sense single-stranded RNA virus. It is contagious in humans and is the cause of the ongoing pandemic of coronavirus disease 2019 (COVID-19) that has been designated a Public Health Emergency of International Concern by the World Health Organization (WHO).
SARS-CoV-2 has close genetic similarity to bat coronaviruses, from which it likely originated. An intermediate reservoir such as a pangolin is also thought to be involved in its introduction to humans. From a taxonomic perspective, SARS-CoV-2 is classified as a strain of the species severe acute respiratory syndrome-related coronavirus (SARSr-CoV).
Because the strain was first discovered in Wuhan, China, it has sometimes been referred to as the "Wuhan virus" or "Wuhan coronavirus", although the World Health Organization (WHO) discourages the use of names based upon locations. To avoid confusion with the disease SARS, the WHO sometimes refers to the virus as "the virus responsible for COVID-19" or "the COVID-19 virus" in public health communications. Both the virus and the disease are often called "coronavirus" by the general public, but scientists and most journalists typically use more precise terms.
Virology
Infection
Human-to-human transmission of SARS-CoV-2 has been confirmed during the 2019–20 coronavirus pandemic. Transmission occurs primarily via respiratory droplets from coughs and sneezes within a range of about 6 feet (1.8 m). Indirect contact via contaminated surfaces is another possible cause of infection. Preliminary research indicates that the virus may remain viable on plastic and steel for up to three days, but does not survive on cardboard for more than one day or on copper for more than four hours. Viral RNA has also been found in stool samples from infected patients.
It is unclear whether the virus is infectious during the incubation period. On 1 February 2020, the World Health Organization (WHO) indicated that "transmission from asymptomatic cases is likely not a major driver of transmission". Thus, most infections in humans are believed to be the result of transmission from subjects exhibiting symptoms of coronavirus disease 2019. However, an epidemiological model of the beginning of the outbreak in China suggested that "pre-symptomatic shedding may be typical among documented infections."
Research from the US NIAID suggested that respiratory droplets with the virus remain viable as an aerosol for at least three hours. On plastic and stainless steel the virus remained detectable after three days; on copper, it took four hours for the virus to lose viability, while on cardboard the virus would be no longer viable after 24 hours. The particles were have found to have a half-life of 66 minutes in an aerosol droplet. Half-lives on different materials were: 5 hours and 38 minutes on stainless steel, 6 hours and 49 minutes on plastic, 3.5 hours on cardboard, and 46 minutes on copper, although the researchers cautioned that there was high variance.
Reservoir
The first known infections from the SARS-CoV-2 strain were discovered in Wuhan, China. The original source of viral transmission to humans remains unclear. However, research into the natural reservoir of the virus strain that caused the 2002–2004 SARS outbreak has resulted in the discovery of many SARS-like bat coronaviruses, most originating in the Rhinolophus genus of horseshoe bats. Two viral nucleic acid sequences found in samples taken from Rhinolophus sinicus show a resemblance of 80% to SARS-CoV-2. A third viral nucleic acid sequence from Rhinolophus affinis collected in Yunnan province has a 96% resemblance to SARS-CoV-2. The WHO considers bats the most likely natural reservoir of SARS-CoV-2, but differences between the bat coronavirus and SARS-CoV-2 suggest that humans were infected via an intermediate host.
A metagenomic study published in 2019 previously revealed that SARS-CoV, the strain of the virus that causes SARS, was the most widely distributed coronavirus among a sample of Sunda pangolins. On 7 February 2020, it was announced that researchers from Guangzhou had discovered a pangolin sample with a viral nucleic acid sequence "99% identical" to SARS-CoV-2. When released, the results clarified that "the receptor-binding domain of the S protein of the newly discovered Pangolin-CoV is virtually identical to that of 2019-nCoV, with one amino acid difference." Pangolins are protected under Chinese law, but their poaching and trading for use in traditional Chinese medicine remains common.
Microbiologists and geneticists in Texas have independently found evidence of reassortment in coronaviruses suggesting involvement of pangolins in the origin of SARS-CoV-2. However, pangolin coronaviruses found to date only share at most 92% of their whole genomes with SARS-CoV-2, which is insufficient to prove pangolins to be the intermediate host; in comparison, the SARS virus responsible for the 2002–2004 outbreak shared 99.8% of its genome with a known civet coronavirus.
Phylogenetics and taxonomy
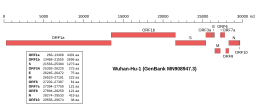 Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2 Genomic organisation of isolate Wuhan-Hu-1, the earliest sequenced sample of SARS-CoV-2 | |
| NCBI genome ID | MN908947 |
|---|---|
| Genome size | 29,903 bases |
| Year of completion | 2020 |
SARS-CoV-2 belongs to the broad family of viruses known as coronaviruses. It is a positive-sense single-stranded RNA (+ssRNA) virus. Other coronaviruses are capable of causing illnesses ranging from the common cold to more severe diseases such as Middle East respiratory syndrome (MERS). It is the seventh known coronavirus to infect people, after 229E, NL63, OC43, HKU1, MERS-CoV, and the original SARS-CoV.
Like the SARS-related coronavirus strain implicated in the 2003 SARS outbreak, SARS-CoV-2 is a member of the subgenus Sarbecovirus (beta-CoV lineage B). Its RNA sequence is approximately 30,000 bases in length. SARS-CoV-2 is unique among known betacoronaviruses in its incorporation of a polybasic cleavage site, a characteristic known to increase pathogenicity and transmissibility in other viruses.
With a sufficient number of sequenced genomes, it is possible to reconstruct a phylogenetic tree of the mutation history of a family of viruses. By 12 January 2020, five genomes of SARS-CoV-2 had been isolated from Wuhan and reported by the Chinese Center for Disease Control and Prevention (CCDC) and other institutions; the number of genomes increased to 42 by 30 January 2020. A phylogenetic analysis of those samples showed they were "highly related with at most seven mutations relative to a common ancestor", implying that the first human infection occurred in November or December 2019. As of 13 March 2020, 410 SARS-CoV-2 genomes sampled on five continents were publicly available.
On 11 February 2020, the International Committee on Taxonomy of Viruses (ICTV) announced that according to existing rules that compute hierarchical relationships among coronaviruses on the basis of five conserved sequences of nucleic acids, the differences between what was then called 2019-nCoV and the virus strain from the 2003 SARS outbreak were insufficient to make them separate viral species. Therefore, they identified 2019-nCoV as a strain of severe acute respiratory syndrome-related coronavirus.
Structural biology
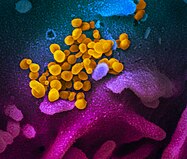
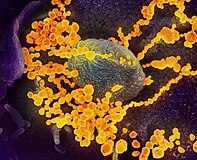 Digitally colourised electron micrographs of SARS-CoV-2 (yellow) emerging from human cells cultured in a laboratory
Digitally colourised electron micrographs of SARS-CoV-2 (yellow) emerging from human cells cultured in a laboratory
Each SARS-CoV-2 virion is approximately 50–200 nanometres in diameter. Like other coronaviruses, SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The spike protein, which has been imaged at the atomic level using cryogenic electron microscopy, is the protein responsible for allowing the virus to attach to the membrane of a host cell.
Protein modeling experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the angiotensin converting enzyme 2 (ACE2) receptors of human cells to use them as a mechanism of cell entry. By 22 January 2020, a group in China working with the full virus genome and a group in the United States using reverse genetics methods independently and experimentally demonstrated that ACE2 could act as the receptor for SARS-CoV-2. Studies have shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain. SARS-CoV-2 may also use basigin to gain cell entry.
Initial spike protein priming by transmembrane protease, serine 2 (TMPRSS2) is essential for entry of SARS-CoV-2. After a SARS-CoV-2 virion attaches to a target cell, the cell's protease TMPRSS2 cuts open the spike protein of the virus, exposing a fusion peptide. The virion then releases RNA into the cell, forcing the cell to produce copies of the virus that are disseminated to infect more cells. SARS-CoV-2 produces at least three virulence factors that promote shedding of new virions from host cells and inhibit immune response.
Epidemiology
Main articles: 2019–20 coronavirus pandemic and COVID-19 testing
Based upon the low variability exhibited among known SARS-CoV-2 genomic sequences, the strain is thought to have been detected by health authorities within weeks of its emergence among the human population in late 2019. The earliest case of infection currently known is thought to have been found on 17 November 2019. The virus subsequently spread to all provinces of China and to more than one hundred other countries in Asia, Europe, North America, South America, Africa, and Oceania. Human-to-human transmission of the virus has been confirmed in all of these regions. On 30 January 2020, SARS-CoV-2 was designated a Public Health Emergency of International Concern by the WHO, and on 11 March 2020 the WHO declared it a pandemic.
As of 10 March 2023 (13:21 UTC), there were 676,609,955Template:Edit sup confirmed cases of infection, of which were in mainland China. While the proportion of infections that result in confirmed infection or progress to diagnosable disease remains unclear, one mathematical model estimated the number of people infected in Wuhan alone at 75,815 as of 25 January 2020, at a time when confirmed infections were far lower. The total number of deaths attributed to the virus was 6,881,955Template:Edit sup as of 10 March 2023 (13:21 UTC); people had recovered from infection by that time. Over a third of all deaths have occurred in Hubei province, where Wuhan is located. Before 24 February 2020, the proportion was over 95%.
The basic reproduction number () of the virus has been estimated to be between 1.4 and 3.9. This means that each infection from the virus is expected to result in 1.4 to 3.9 new infections when no members of the community are immune and no preventive measures are taken.
References
- Jiang S, Shi Z, Shu Y, et al. (19 February 2020). "A distinct new name is needed for the new coronavirus". The Lancet. Retrieved 16 March 2020.
- ^ Gobalenya AE, Baker SC, Baric RS, et al. (March 2020). "The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2". Nature Microbiology: 1–9. doi:10.1038/s41564-020-0695-z. PMID 32123347.
- "Coronavirus disease named Covid-19". BBC News Online. 11 February 2020. Archived from the original on 15 February 2020. Retrieved 15 February 2020.
- World Health Organization (2020). Surveillance case definitions for human infection with novel coronavirus (nCoV): interim guidance v1, January 2020 (Report). World Health Organization. hdl:10665/330376. WHO/2019-nCoV/Surveillance/v2020.1.
- "Healthcare Professionals: Frequently Asked Questions and Answers". United States Centers for Disease Control and Prevention (CDC). 11 February 2020. Archived from the original on 14 February 2020. Retrieved 15 February 2020.
- "About Novel Coronavirus (2019-nCoV)". United States Centers for Disease Control and Prevention (CDC). 11 February 2020. Archived from the original on 11 February 2020. Retrieved 25 February 2020.
- "New-type coronavirus causes pneumonia in Wuhan: expert". Xinhua. Archived from the original on 9 January 2020. Retrieved 9 January 2020.
- ^ "CoV2020". GISAID EpifluDB. Archived from the original on 12 January 2020. Retrieved 12 January 2020.
- ^ Wee SL, McNeil Jr. DG, Hernández JC (30 January 2020). "W.H.O. Declares Global Emergency as Wuhan Coronavirus Spreads". The New York Times. Archived from the original on 30 January 2020. Retrieved 30 January 2020.
- ^ Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. (February 2020). "A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster". The Lancet. 395 (10223): 514–523. doi:10.1016/S0140-6736(20)30154-9. PMID 31986261.
- ^ Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. (February 2020). "A pneumonia outbreak associated with a new coronavirus of probable bat origin". Nature. 579 (7798): 270–273. doi:10.1038/s41586-020-2012-7. PMID 32015507.
- Perlman S (February 2020). "Another Decade, Another Coronavirus". The New England Journal of Medicine. 382 (8): 760–762. doi:10.1056/NEJMe2001126. PMID 31978944.
- ^ Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M (April 2020). "The 2019-new coronavirus epidemic: Evidence for virus evolution". Journal of Medical Virology. 92 (4): 455–459. doi:10.1002/jmv.25688. PMID 31994738.
- World Health Organization (2020). Novel Coronavirus (2019-nCoV): situation report, 22 (Report). World Health Organization. hdl:10665/330991.
- Shield C (7 February 2020). "Coronavirus: From bats to pangolins, how do viruses reach us?". Deutsche Welle. Retrieved 13 March 2020.
- Huang P (22 January 2020). "How Does Wuhan Coronavirus Compare with MERS, SARS and the Common Cold?". NPR. Archived from the original on 2 February 2020. Retrieved 3 February 2020.
- Fox D (2020). "What you need to know about the Wuhan coronavirus". Nature. doi:10.1038/d41586-020-00209-y. ISSN 0028-0836.
- Yam K. "GOP lawmakers continue to use 'Wuhan virus' or 'Chinese coronavirus'". NBC News. Retrieved 12 March 2020.
{{cite news}}: CS1 maint: url-status (link) - Dorman S (11 March 2020). "McCarthy knocks Dems after they claim saying 'Chinese coronavirus' is racist". Fox News. Retrieved 12 March 2020.
{{cite news}}: CS1 maint: url-status (link) - World Health Organization (2015). World Health Organization best practices for the naming of new human infectious diseases (Report). World Health Organization. hdl:10665/163636. WHO/HSE/FOS/15.1.
- "Naming the coronavirus disease (COVID-2019) and the virus that causes it". World Health Organization. Retrieved 24 February 2020.
From a risk communications perspective, using the name SARS can have unintended consequences in terms of creating unnecessary fear for some populations.... For that reason and others, WHO has begun referring to the virus as "the virus responsible for COVID-19" or "the COVID-19 virus" when communicating with the public. Neither of these designations are intended as replacements for the official name of the virus as agreed by the ICTV.
- Harmon A (4 March 2020). "We Spoke to Six Americans with Coronavirus". New York Times. Retrieved 16 March 2020.
- Edwards E (25 January 2020). "How does coronavirus spread?". NBC News. Archived from the original on 28 January 2020. Retrieved 13 March 2020.
- "How COVID-19 Spreads". U.S. Centers for Disease Control and Prevention (CDC). 27 January 2020. Archived from the original on 28 January 2020. Retrieved 29 January 2020.
- "Getting your workplace ready for COVID-19" (PDF). World Health Organization. 27 February 2020. Retrieved 3 March 2020.
{{cite web}}: CS1 maint: url-status (link) - van Doremalen N, Bushmaker T, Morris DH, et al. (2020). "Correspondence: Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1". The New England Journal of Medicine. doi:10.1056/NEJMc2004973. PMID 32182409.
- Holshue ML, DeBolt C, Lindquist S, et al. (March 2020). "First Case of 2019 Novel Coronavirus in the United States". The New England Journal of Medicine. 382 (10): 929–936. doi:10.1056/NEJMoa2001191. PMID 32004427.
- Kupferschmidt K (February 2020). "Study claiming new coronavirus can be transmitted by people without symptoms was flawed". Science. doi:10.1126/science.abb1524.
- World Health Organization (2020). Novel Coronavirus (2019-nCoV): situation report, 12 (Report). World Health Organization. hdl:10665/330777.
- Li R, Pei S, Chen B, et al. (16 March 2020). "Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2)". Science: eabb3221. doi:10.1126/science.abb3221. Retrieved 17 March 2020.
- Gene Emery (17 March 2020). "Coronavirus can persist in air for hours and on surfaces for days: study". Reuters.
- van Doremalen, Neeltje; Morris, Dylan H.; Bushmaker, Trenton; Holbrook, Myndi G.; Gamble, Amandine; Willamson, Brandi N.; Tamin, Azaibi; Harcourt, Jennifer L.; Thornberg, Natalie J.; Gerber, Susan I.; Lloyd-Smith, James O.; de Wit, Emmie; Munster, Vincent J. (17 March 2020). "Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1". New England Journal of Medicine. doi:10.1056/NEJMc2004973. ISSN 0028-4793. PMID 32182409. Retrieved 18 March 2020.
- ^ Cohen J (January 2020). "Wuhan seafood market may not be source of novel virus spreading globally". Science. doi:10.1126/science.abb0611. ISSN 0036-8075.
- Eschner K (28 January 2020). "We're still not sure where the Wuhan coronavirus really came from". Popular Science. Archived from the original on 29 January 2020. Retrieved 30 January 2020.
- Yu WB, Tang GD, Zhang L, Corlett RT (21 February 2020). "Decoding evolution and transmissions of novel pneumonia coronavirus using the whole genomic data" (Document). doi:10.12074/202002.00033 (inactive 26 February 2020).
{{cite document}}: Cite document requires|publisher=(help); Unknown parameter|url=ignored (help)CS1 maint: DOI inactive as of February 2020 (link) - "Bat SARS-like coronavirus isolate bat-SL-CoVZC45, complete genome" (Document). National Center for Biotechnology Information (NCBI). 15 February 2020.
{{cite document}}: Unknown parameter|access-date=ignored (help); Unknown parameter|url=ignored (help) - "Bat SARS-like coronavirus isolate bat-SL-CoVZXC21, complete genome" (Document). National Center for Biotechnology Information (NCBI). 15 February 2020.
{{cite document}}: Unknown parameter|access-date=ignored (help); Unknown parameter|url=ignored (help) - "Bat coronavirus isolate RaTG13, complete genome" (Document). National Center for Biotechnology Information (NCBI). 10 February 2020.
{{cite document}}: Unknown parameter|access-date=ignored (help); Unknown parameter|url=ignored (help) - "Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)" (Document). World Health Organization (WHO). 24 February 2020.
{{cite document}}: Unknown parameter|access-date=ignored (help); Unknown parameter|url=ignored (help) - ^ Cyranoski D (26 February 2020). "Mystery deepens over animal source of coronavirus". Nature. 579 (7797): 18–19. doi:10.1038/d41586-020-00548-w. PMID 32127703.
- Liu P, Chen W, Chen JP (October 2019). "Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins (Manis javanica)". Viruses. 11 (11): 979. doi:10.3390/v11110979. PMC 6893680. PMID 31652964.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Cyranoski D (7 February 2020). "Did pangolins spread the China coronavirus to people?". Nature. doi:10.1038/d41586-020-00364-2.
- Xiao K, Zhai J, Feng Y (February 2020). "Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins". bioRxiv (preprint). doi:10.1101/2020.02.17.951335.
- Kelly, Guy (1 January 2015). "Pangolins: 13 facts about the world's most hunted animal". The Telegraph. Retrieved 9 March 2020.
- Wong MC, Cregeen SJ, Ajami NJ, Petrosino JF (February 2020). "Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019". bioRxiv (preprint). doi:10.1101/2020.02.07.939207.
- Zhu N, Zhang D, Wang W, et al. (February 2020). "A Novel Coronavirus from Patients with Pneumonia in China, 2019". The New England Journal of Medicine. 382 (8): 727–733. doi:10.1056/NEJMoa2001017. PMID 31978945.
- Hui DS, I Azhar E, Madani TA, et al. (February 2020). "The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — The latest 2019 novel coronavirus outbreak in Wuhan, China". International Journal of Infectious Diseases. 91: 264–266. doi:10.1016/j.ijid.2020.01.009. PMID 31953166. [REDACTED]
- "Phylogeny of SARS-like betacoronaviruses". nextstrain. Archived from the original on 20 January 2020. Retrieved 18 January 2020.
- Wong AC, Li X, Lau SK, Woo PC (February 2019). "Global Epidemiology of Bat Coronaviruses". Viruses. 11 (2): 174. doi:10.3390/v11020174. PMC 6409556. PMID 30791586.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Walls AC, Park YJ, Tortorici MA, et al. (9 March 2020). "Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein". Cell. doi:10.1016/j.cell.2020.02.058. PMID 32155444.
- Andersen KG, Rambaut A, Lipkin WI, et al. (16 February 2020). "The Proximal Origin of SARS-CoV-2". Virological. Retrieved 4 March 2020.
- Coutard B, Valle C, de Lamballerie X, et al. (February 2020). "The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade". Antiviral Research. 176: 104742. doi:10.1016/j.antiviral.2020.104742. PMID 32057769.
- "Initial genome release of novel coronavirus". Virological. 11 January 2020. Archived from the original on 12 January 2020. Retrieved 12 January 2020.
- ^ Bedford T, Neher R, Hadfield N, et al. "Genomic analysis of nCoV spread: Situation report 2020-01-30". nextstrain.org. Retrieved 18 March 2020.
- Hodcroft E, Müller N, Wagner C, et al. "Genomic analysis of COVID-19 spread: Situation report 2020-03-13". nextstrain.org. Retrieved 18 March 2020.
- Chen N, Zhou M, Dong X, et al. (15 February 2020). "Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study". The Lancet. 395 (10223): 507–513. doi:10.1016/S0140-6736(20)30211-7. PMID 32007143.
- ^ Wu C, Liu Y, Yang Y, et al. (February 2020). "Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods". Acta Pharmaceutica Sinica B. doi:10.1016/j.apsb.2020.02.008.
- ^ Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. (February 2020). "Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation". Science. 367 (6483): 1260–1263. doi:10.1126/science.abb2507. PMID 32075877.
- Mandelbaum RF (19 February 2020). "Scientists Create Atomic-Level Image of the New Coronavirus's Potential Achilles Heel". Gizmodo. Retrieved 13 March 2020.
- Xu X, Chen P, Wang J, et al. (March 2020). "Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission". Science China Life Sciences. 63 (3): 457–460. doi:10.1007/s11427-020-1637-5. PMID 32009228.
- Letko M, Munster V (January 2020). "Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV". bioRxiv (preprint). doi:10.1101/2020.01.22.915660.
- Letko M, Marzi A, Munster V (February 2020). "Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses". Nature Microbiology. doi:10.1038/s41564-020-0688-y. PMID 32094589.
- El Sahly HM. "Genomic Characterization of the 2019 Novel Coronavirus". The New England Journal of Medicine. Retrieved 9 February 2020.
- Gralinski LE, Menachery VD (January 2020). "Return of the Coronavirus: 2019-nCoV". Viruses. 12 (2): 135. doi:10.3390/v12020135. PMID 31991541.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. (February 2020). "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding". The Lancet. 395 (10224): 565–574. doi:10.1016/S0140-6736(20)30251-8. PMID 32007145.
- Wang K, Chen W, Zhou YS, et al. (2020). "SARS-CoV-2 invades host cells via a novel route: CD147-spike protein". bioRxiv (preprint). doi:10.1101/2020.03.14.988345.
- Hoffman M, Kliene-Weber H, Krüger N, et al. (16 April 2020). "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor". Cell. 181: 1–10. doi:10.1016/j.cell.2020.02.052. PMID 32142651.
- "Anatomy of a Killer: Understanding SARS-CoV-2 and the drugs that might lessen its power". The Economist. 12 March 2020. Retrieved 14 March 2020.
- Oberholzer M, Febbo P (19 February 2020). "What We Know Today about Coronavirus SARS-CoV-2 and Where Do We Go from Here". Genetic Engineering and Biotechnology News. Retrieved 13 March 2020.
- Ma J (13 March 2020). "Coronavirus: China's first confirmed Covid-19 case traced back to November 17". South China Morning Post. Retrieved 16 March 2020.
- ^ "COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)". ArcGIS. Johns Hopkins University. Retrieved 10 March 2023.
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. (March 2020). "Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany". The New England Journal of Medicine. 382 (10): 970–971. doi:10.1056/NEJMc2001468. PMID 32003551.
- Molteni M (30 January 2020). "The Coronavirus Is Now Infecting More People Outside China". Wired. Retrieved 13 March 2020.
- Khalik S (4 February 2020). "Coronavirus: Singapore reports first cases of local transmission; 4 out of 6 new cases did not travel to China". The Straits Times. Archived from the original on 4 February 2020. Retrieved 5 February 2020.
- "Ecuador confirms five new cases of coronavirus, all close to initial patient". Reuters. 2 March 2020. Retrieved 5 March 2020.
- "Algeria confirms two more coronavirus cases". Reuters. 2 March 2020. Retrieved 5 March 2020.
- "Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV)". World Health Organization (WHO) (Press release). 30 January 2020. Archived from the original on 31 January 2020. Retrieved 30 January 2020.
- McKay B, Calfas J, Ansari T (11 March 2020). "Coronavirus Declared Pandemic by World Health Organization". The Wall Street Journal. Retrieved 12 March 2020.
{{cite news}}: CS1 maint: url-status (link) - "WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020". World Health Organization (WHO) (Press release). 11 March 2020. Retrieved 12 March 2020.
{{cite press release}}: CS1 maint: url-status (link) - Branswell H (30 January 2020). "Limited data on coronavirus may be skewing assumptions about severity". STAT. Archived from the original on 1 February 2020. Retrieved 13 March 2020.
- Wu JT, Leung K, Leung GM (February 2020). "Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study". The Lancet. 395 (10225): 689–697. doi:10.1016/S0140-6736(20)30260-9. PMID 32014114.
- Boseley S, McCurry J (30 January 2020). "Coronavirus deaths leap in China as countries struggle to evacuate citizens". The Guardian. Retrieved 10 March 2020.
{{cite news}}: CS1 maint: url-status (link) - Paulinus A (25 February 2020). "Coronavirus: China to repay Africa in safeguarding public health". The Sun. Retrieved 10 March 2020.
{{cite news}}: CS1 maint: url-status (link) - Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. (January 2020). "Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia". The New England Journal of Medicine. doi:10.1056/NEJMoa2001316. PMID 31995857.
- Riou J, Althaus CL (January 2020). "Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020". Euro Surveillance. 25 (4). doi:10.2807/1560-7917.ES.2020.25.4.2000058. PMC 7001239. PMID 32019669.
Further reading
- Brüssow H (March 2020). "The Novel Coronavirus – A Snapshot of Current Knowledge". Microbial Biotechnology. 2020: 1–6. doi:10.1111/1751-7915.13557. PMID 32144890.
- Habibzadeh P, Stoneman EK (February 2020). "The Novel Coronavirus: A Bird's Eye View". The International Journal of Occupational and Environmental Medicine. 11 (2): 65–71. doi:10.15171/ijoem.2020.1921. PMID 32020915.
- World Health Organization (2 March 2020). Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020 (Report). World Health Organization. hdl:10665/331329. WHO/COVID-19/laboratory/2020.4. License: CC BY-NC-SA 3.0.
- World Health Organization (2020). WHO R&D Blueprint: informal consultation on prioritization of candidate therapeutic agents for use in novel coronavirus 2019 infection, Geneva, Switzerland, 24 January 2020 (Report). World Health Organization. hdl:10665/330680. WHO/HEO/R&D Blueprint (nCoV)/2020.1.
External links
[REDACTED] Scholia has a profile for SARS-CoV-2 (Q82069695).- "Coronavirus Disease 2019 (COVID-19)". Centers for Disease Control and Prevention (CDC).
- "Coronavirus disease (COVID-19) outbreak". World Health Organization (WHO).
- "SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) Sequences". National Center for Biotechnology Information (NCBI).
- "COVID-19 Resource Centre". The Lancet.
- "Coronavirus (Covid-19)". The New England Journal of Medicine.
- "Covid-19: Novel Coronavirus Content Free to Access". Wiley.
- "2019-nCoV Data Portal". Virus Pathogen Database and Analysis Resource.
| Classification | D |
|---|
| COVID-19 pandemic | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon identifiers | |
|---|---|
| Severe acute respiratory syndrome-related coronavirus | |
 ) of the virus has been estimated to be between 1.4 and 3.9. This means that each infection from the virus is expected to result in 1.4 to 3.9 new infections when no members of the community are
) of the virus has been estimated to be between 1.4 and 3.9. This means that each infection from the virus is expected to result in 1.4 to 3.9 new infections when no members of the community are